Data Analytics
Genetic data analytic solutions for managing large volumes of genomic data for bioinformaticians, researchers and scientists.
Bioinformatics and data science solutions
Overview
Our laboratory services generate a very large amount of data. With nearly 3 million whole genome profiles generated by our technologies, each with potentially millions of markers produced on average by DArTreseq, we store over 100 Tb of sequence data.
The full value of this data is unlocked through our highly developed skills in data analytics. Using a wide range of tools, we are able to offer analytical services, including our software solutions.
Data analytics software platform
Our platform is a comprehensive and scalable solution suite that supports breeders through all stages of:
- Data collection
- Quality control
- Prediction modelling
- Decision support
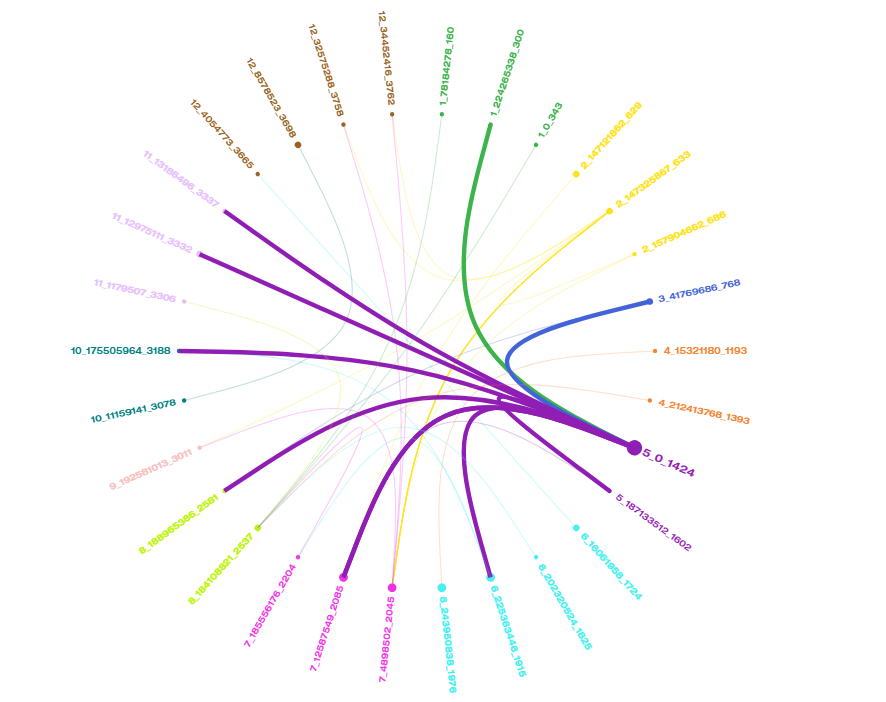
Markers with significant correlations in Capsicum
1 of 1Innovation and R&D
Access to ground-breaking innovations to improve detection and evaluation of both genetic and environmental factors.
Predictive modelling
Predictive modelling and DArT’s Universal Test Bench (UTB) to assist in the selection of the right algorithm for any particular data set.
GEMIA
GEMIA – Genomics, Environment & Management Interaction Analytics – which harnesses whole-of-data modelling, computation, and parallel processing for multi-way interactions, via associated traits of interest.
There are two domains in our data analytics:
- Support for our genome profiling services at various levels of marker density which focuses on effective extraction of marker information from the sequences; and
- Support for downstream applications of marker data produced by our DNA profiling services. Both of these areas are enabled by the KDCompute application – an integral part of our KDDart platform
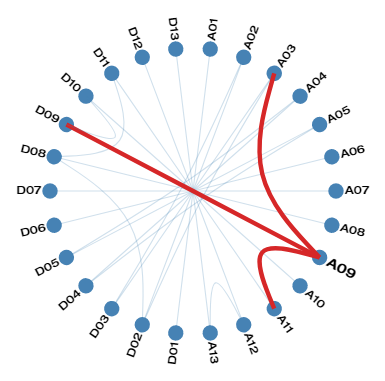
Linkage Disequilibrium among markers on separate chromosomes in Cotton
1 of 1KDCompute
KDCompute can operate as a standalone application or in connection with a database. When connected to DArTdb via DArTsoft14 plugin KDCompute enables rapid marker data extraction from sequences produced on NGS platforms. Tens of thousands of libraries/samples can be effectively processed within hours; and in addition to marker data, a range of metadata for markers are also extracted and reported.
KDCompute can also connect to KDDart database and perform a range of analysis relevant to breeding or ecology; including experimental design, analysis of field data, population genetics, marker trait associations (GWAS) and Genomic Selection. Dozens of algorithms of various levels of complexity are “wrapped” into simple user interfaces which make the results just a few clicks away.
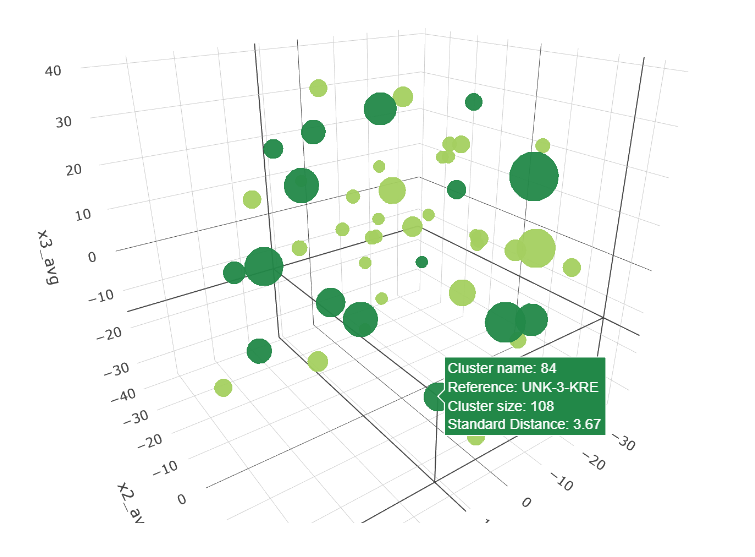
Clustering varieties of Sweet Potatoes
1 of 1